Artificial Intelligence Ultrafast High-quality Nuclear Magnetic Resonance Spectroscopy Reconstruction
Nuclear magnetic resonance (NMR) spectroscopy is an invaluable biophysical tool in modern chemistry and life sciences. However, duration of NMR experiments increases rapidly with spectral resolution and dimensionality, which often imposes unbearable limitations due to low sample stability and/or excessive costs of NMR measurement time. To accelerate the data acquisition and optimize sensitivity, modern NMR experiments are often acquired using the Non-Uniform Sampling (NUS) approach, where only a small fraction of traditional NMR measurements, usually called free induction decay (FID), is performed. Inevitably, a spectra reconstruction method is most needed to reliably and quickly repair the information loss caused by undersampling.
This work is the proof-of-concept of application of deep learning (DL), an artificial intelligence technique, for high-quality, reliable, and very fast NMR spectra reconstruction from limited experimental data (X. Qu et al., arXiv:1904.05168v1, 9 Apr 2019.;X. Qu et al., Angew. Chem. -Int. Edit., 59(26):10297-10300, 2020).
[Figure 1]. Deep Learning-based Fast Biomedical Nuclear Magnetic Resonance Spectroscopy.
Based on recent developments in the field of signal processing theory and the exponential characteristics of NMR signals, a deep learning method utilizing convolutional neural network (CNN) is designed. It can be successfully trained using solely synthetic NMR data, which lifts the prohibiting demand for a large volume of realistic training data usually required in the deep learning approach. Spectrum aliasing artifacts introduced by NUS data are gradually removed with five consecutive dense CNN blocks with data consistency constrained to the sampled data points.

[Figure 2]. The architectures of NMR spectra reconstruction with deep learning.
The proposed DL method can fastly reconstruct the high-quality NMR spectra of small, large and disordered proteins from NUS data. The reconstruction performance by DL is comparable with the state-of-the-art iterative methods low-rank (LR) and compressed sensing (CS),but it shows outstanding advantage in computational time: 5~8% of LRHM for 2D spectra and 12~25% of CS for 3D spectra.
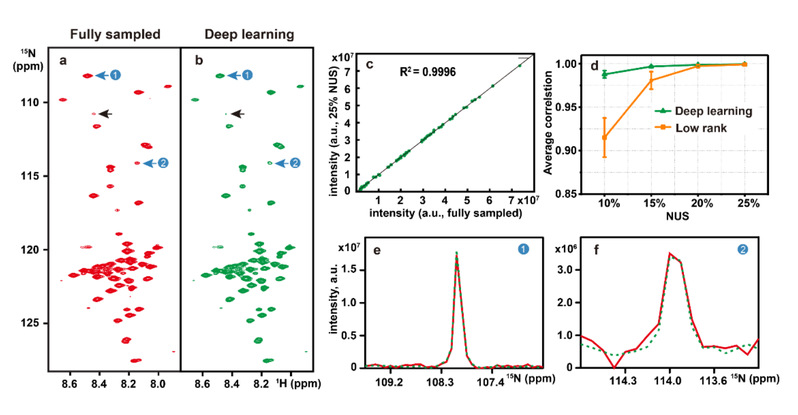
[Figure 3]. Reconstruction of a 2D 1H– 15N HSQC spectrum of the cytosolic domain of CD79b protein from the B-cell receptor. (a) and (b) are the fully sampled spectrum and deep learning NMR reconstruction from 25% NUS data, respectively. (c) Peak intensity correlations between fully sampled spectrum and reconstructed spectrum. (d) denotes the peak intensity correlation obtained with the deep learning and low rank methods under different NUS levels. (e) and (f) are zoomed out 1D 15N traces of (a). Red and green lines represent the reference and the reconstructed spectra, respectively.
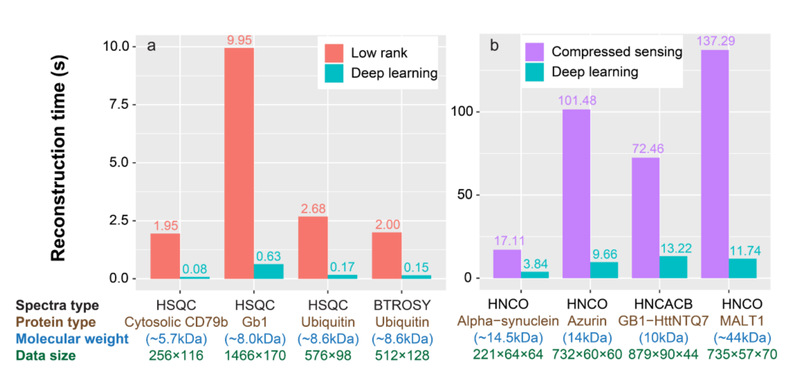
[Figure 4]. Computational time for the reconstructions of (a) 2D spectra and (b) 3D spectra with low rank, compressed sensing and deep learning. Note: The listed below each bar are spectra type, the corresponding protein, molecular weight, and spectra size, respectively. For 2D (3D) spectra size of the directly detected dimension is followed by size(s) of the indirect dimension(s).
After publishing the paper, our team was invited by the top journal to write a review about “deep learning magnetic resonance spectroscopy” and published it (D. Chen et al., Chem. -Eur. J., DOI: 10.1002/chem.202000246, 2020.). It is worth mentioning that this method has been granted a China National Patent and has been industrialized by Q. One Instruments Ltd. In the foreseeable future, it is expected to expand to higher-dimensional data reconstruction and different NMR experiments, such as diffusion spectroscopy.